StainSTORM started from a simple idea: if we want spatial biology workflows to be truly reproducible and scalable at different lab sites, the “lab” itself has to become something we can deploy, version, and operate like software in a very distributed manner. The project (or rather multiple projects such as the TAB-funded RunFAST and the MDC-funded Gateway Project) brings together two largely hardware-matched automation stations, one in Helsinki and one in Jena, and connects them through a shared orchestration and analysis backbone using Arkitekt. What makes it special is not just the technical stack, but the way the collaboration happened: the system was developed remotely across multiple institutions and then brought to life in a focused on-site commissioning effort. This was done in a seris of online meetings and very intensive and even more fun hackathons.

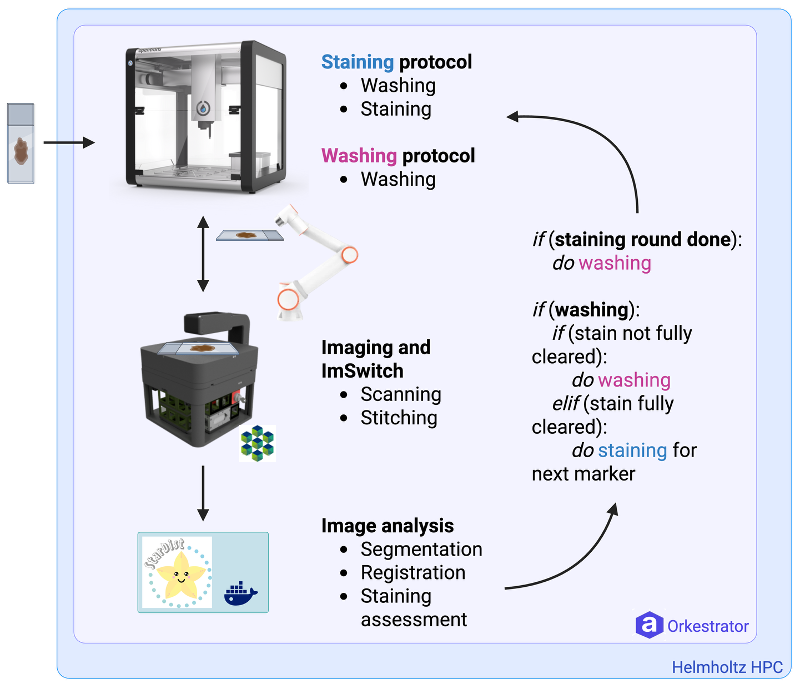
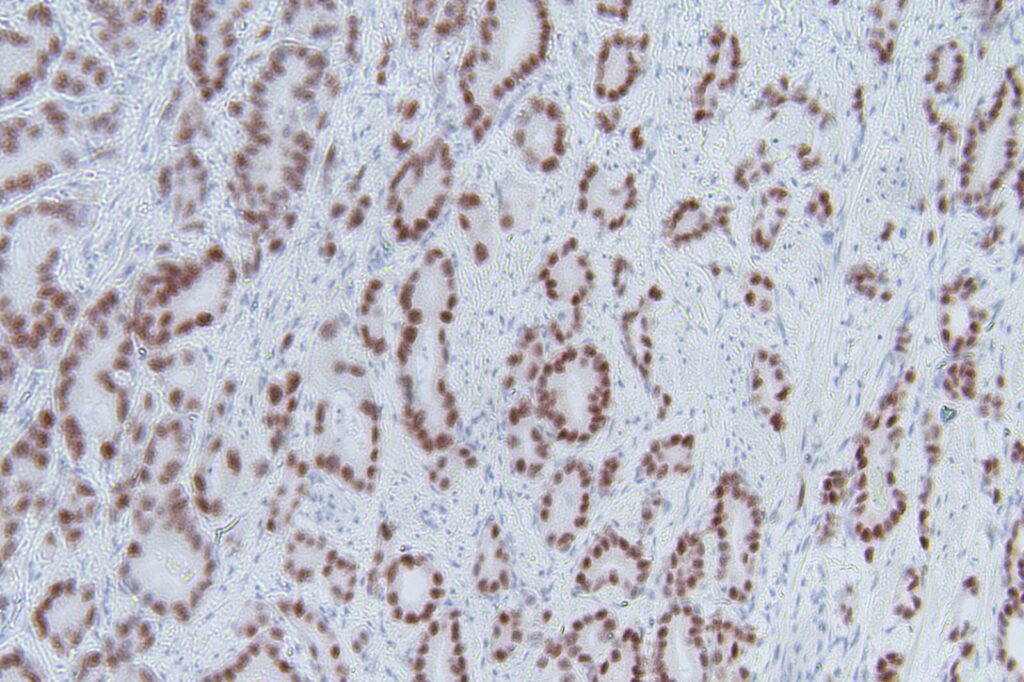
At the center of the workflow is a cyclic chromogenic immunohistochemistry protocol designed for FFPE tissue slides. The goal is to run repeated staining rounds on the same sample, image each round, and use the acquired data for both documentation and quality control inside the loop. That requires precise and repeatable liquid handling, reliable sample logistics between instruments, robust imaging, and an analysis pipeline that can keep up with the data volume and provide actionable readouts.
The University of Helsinki team provided the biological and experimental anchor of the project. They drove the wet-lab requirements, shaped the staining protocol around real research constraints, and ensured that what we automate is not just a “demo pipeline” but something aligned with practical tissue workflows. The workflow using the Opentrons OT2 was already part of their daily routine, but it lagged high-throughput. Their perspective defined the success criteria: the staining needs to be consistent across cycles, the images need to be interpretable for downstream analysis, and the system has to be robust enough to run for extended periods without constant supervision.
The automation layer is built around the idea that each device should expose a hardware agnostic service that is reliable and contains high-level actions that can be composed into a larger workflow. The Opentrons OT2 plays a key role here by executing the repeated staining and washing steps in a controlled, repeatable manner. The Fairino FR5 robotic arm layer adds what is often missing in lab automation: physical integration for moving slides from a-to-bo. With the Fairino arm integration, the system can transport slide racks between stations and place them reproducibly for imaging, enabling continuous operation and tighter coupling between wet lab steps and imaging-based checks. To make this hole step simple, we designed a custom sample holder ready to hold multiple slides for sequential staining in the opentrons and then moving it back and forth to the microscope:

OpenUC2’s core contribution is (obviously) the microscope. We provided the OpenUC2 FRAME microscope as an affordable, modular imaging node that can be tightly integrated into an automation stack. Beyond the hardware, the microscope is designed to operate headless and to expose scanning functions that fit into a service-based architecture. That matters because the microscope is not treated as a special one-off instrument, but as a programmable component in a distributed workflow. The OpenUC2 microscope captures tile scans that can be stitched into large mosaics and compared across staining cycles, making it possible to support quality control and downstream segmentation without relying on proprietary scanning infrastructure. We integrated the necessary services through arkitekt in a simple controller: https://github.com/openUC2/ImSwitch/blob/master/imswitch/imcontrol/controller/controllers/ArkitektController.py. this one adapts the function to scan a slide to the Service world of Arkitekt.
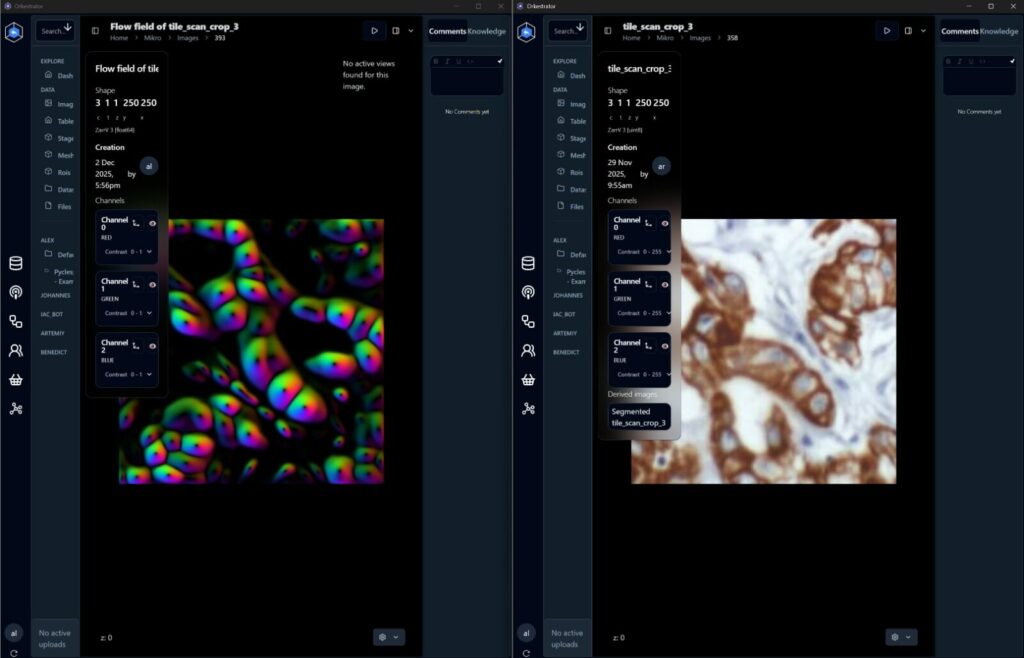
The compute and analysis side is where the workflow becomes more than just automation. The Max Delbruck Center and the Helmholtz HPC environment provide the computational home for large-scale image processing: stitching, registration across cycles, segmentation, and exploratory biomarker-driven analysis. This is essential because cyclic workflows quickly generate many gigabytes of image data, and turning that into per-cell and per-region measurements requires scalable resources. Equally important is the idea of traceability: analysis is part of the experiment, not an afterthought, and it needs to be reproducible alongside the wet-lab steps.
Arkitekt ties everything together. Instead of building a monolithic control program, we structured StainSTORM as a set of small services, each running in containers, often on embedded nodes such as Raspberry Pi computers. The orchestrator composes these services into a step-chain workflow that behaves like a state machine, with explicit transitions and decision points. This architecture also enabled the remote-first development process: we could define the workflow, test it against mock services, swap in real hardware drivers once they were ready, and then deploy a consistent stack to both sites.

What emerged from this partnership is a blueprint for how cross-lab automation can work in practice. Each partner contributed what they are best at—biological requirements and validation, robotic handling and protocol execution, open and modular microscopy, scalable compute and analysis, and the orchestration framework that turns it into one coherent system. The result is not just an automated staining pipeline, but a reproducible way to build and share complex lab workflows across geography, budgets, and infrastructure levels.
This project was and still is funded from multiple sources. We contributed to a Helmholtz Gateway Project at the Helmholtz MDC through the TP Image Data Analysis Kooperation HPC Gateway (28/2025 KSCH – MDC / OpenUC2). We also thank the Thüringer Aufbaubank (TAB) for their financial support through the RunFAST project supported the the EU Council EFRE.


